
Order the NovaSeq X Series
Advanced chemistry, optics, and informatics combine to deliver exceptional speed and data quality, outstanding throughput and scalability.
A library prep and enrichment kit that enables sensitive, culture-free detection and analysis of common and underrecognized uropathogens and AMR genes.
Assay time
Hands-on time
Input quantity
The Urinary Pathogen ID/AMR Panel (UPIP) is a next-generation sequencing (NGS) research panel that enables detection and quantification of over 170 uropathogens responsible for urinary tract infections (UTIs) and co-infections and covers more than 3700 markers and their alleles associated with antimicrobial resistance (AMR).
The Urinary Pathogen ID/AMR Enrichment Panel Kit contains all library prep and enrichment reagents, indexes, and panel probes for on-bead tagmentation followed by a single hybridization step for generating enriched libraries. Integration of the enrichment kit with DNA extraction, sequencing, and Explify UPIP Data Analysis allows a comprehensive and simple workflow for detection and identification of UTIs.
| Assay time | < 9 hr library prep and enrichment time |
|---|---|
| Automation capability | Liquid Handling Robots |
| Content specifications |
Detects 120+ bacteria, 14 fungi, 35 viruses, 4 parasites, including 27 sexually transmitted pathogens, and > 3700 antimicrobial resistance markers |
| Description |
A streamlined workflow that incorporates DNA extraction, library preparation using the Urinary Pathogen ID/AMR Enrichment Kit, sequencing, and Explify UPIP Data Analysis for detection and identification of uropathogens and antimicrobial resistance markers from a range of sample types. |
| Hands-on time | < 2 hr library prep and enrichment time |
| Input quantity | Based on volume (not concentration dependent) |
| Instruments | MiSeq System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiSeqDx in Research Mode, MiniSeq System, NextSeq 550Dx in Research Mode |
| Mechanism of action | On-bead tagmentation followed by a single hybridization step |
| Method | Target Enrichment, Targeted DNA Sequencing |
| Multiplexing | Up to 384 samples in a single run with unique dual indexes |
| Nucleic acid type | DNA |
| Sample type details |
DNA extracted from human urinary clinical research samples and other surveillance related samples such as wastewater |
| Specialized sample types | Not FFPE-Compatible |
| Species category | Other, Human, Virus, Bacteria |
| Species details |
fungi, parasites |
| Technology | Sequencing |
No accessories needed.
The Urinary Pathogen ID/AMR Enrichment Kit is used for detection and identification of over 170 uropathogens and more than 3700 antimicrobial resistance markers and their alleles from a range of sample types including clinical urinary research, environmental, and other sample types.
Urinary Pathogen ID/AMR Enrichment Kit
As a hypothesis-free method, NGS can distinguish between infectious disease strains that differ by as little as one SNP, and replace multiple tests.
Target enrichment captures genomic regions of interest through hybridization. Learn more about target enrichment and how it can be used to assess multiple samples at once.
Shotgun metagenomic sequencing
Comprehensively sample all genes in all organisms present in a given complex sample to evaluate bacterial diversity and detect unculturable microorganisms.
Library Prep and Array Kit Selector
Find the right sequencing library preparation kit or microarray for your needs. Filter by method, species, and more. Compare, share, and order kits.

Urinary Pathogen ID/AMR Panel Kit can detect and quantify 174 organisms, including common and less common uropathogens and 27 sexually transmitted pathogens.
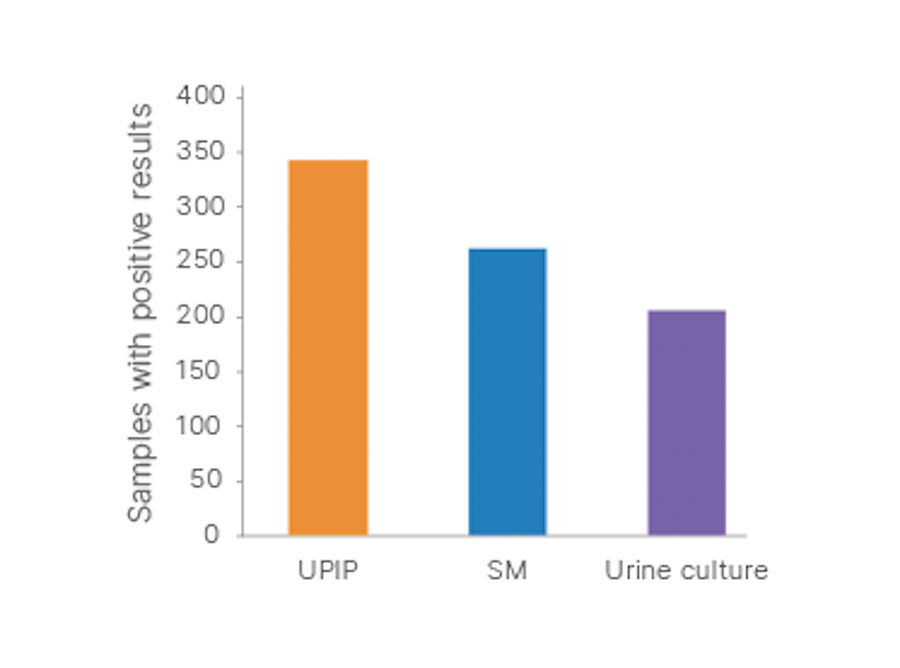

The Urinary Pathogen ID/AMR Panel (UPIP) Kit offers more sensitive pathogen identification than shotgun metagenomics (SM) and culture methods. One or more uropathogens were detected in 342/399 (86%) urine samples by UPIP, in 262/399 samples (66%) by SM, and in 205/399 samples (51%) by urine culture.
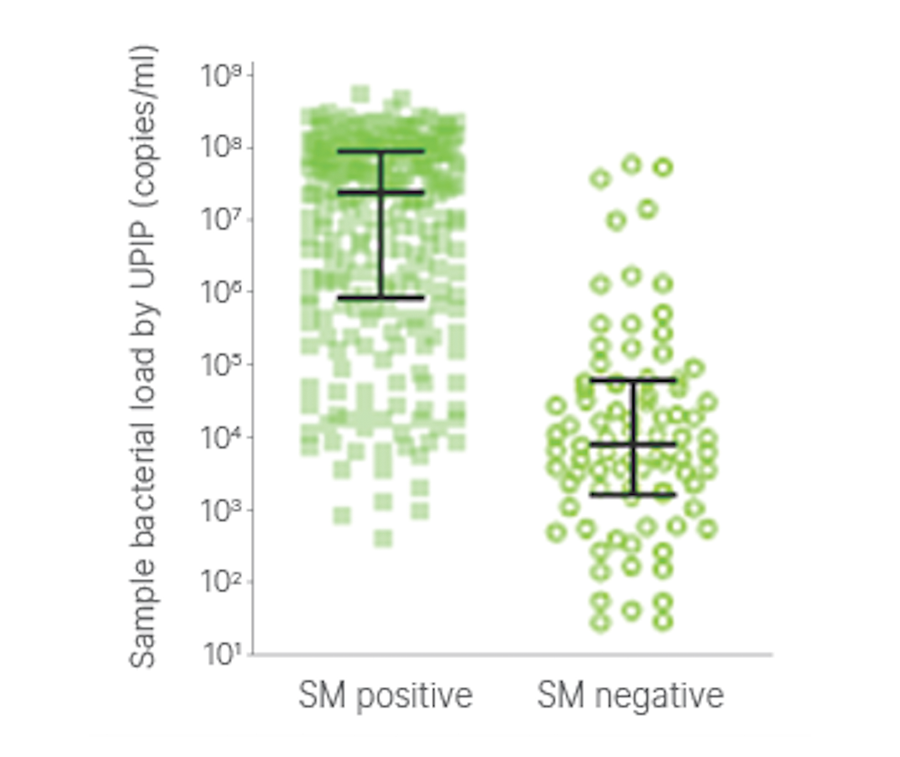

Bacterial load measured by UPIP was higher in SM positive than in SM negative samples (p < 0.0001), consistent with the prediction of improved analytical sensitivity of targeted enrichment sequencing compared with SM. Detection of bacteria in the SM negative samples further demonstrates the improved sensitivity of enrichment compared to shotgun metagenomics.
Urinary Pathogen ID/AMR Enrichment Kit Set A (RUO) (96 indexes,96 samples)
20090308
Includes all reagents necessary for library prep and enrichment including tagementation and PCR reagents, IDT for Illumina DNA/RNA UD Indexes Set A, and Urinary Pathogen ID/AMR Panel to prepare libraries from 96 samples followed by 32, 3-plex enrichment reactions.
List Price:
Discounts:
Urinary Pathogen ID/AMR Enrichment Kit Set B (RUO) (96 indexes, 96 samples)
20090309
The Urinary Pathogen ID/AMR Enrichment Kit (UPIP) contains all library prep reagents, indexes and panel used to target 3,500 antimicrobial resistance (AMR) markers, 121 bacteria, 14 fungi, and 35 viruses and 4 parasites. Coupled with the Illumina DNA prep with Enrichment kit and analysis with the UPIP Explify app on BaseSpace Sequence Hub, the UPIP enrichment panel enables broad range detection of urinary pathogens and profiling of AMR to identify mixed co-infections. Kit contains library prep reagents, indexes (Set B) and panel to prepare 96 samples at 3-plex and analysis with the UPIP Explify app.
List Price:
Discounts:
Urinary Pathogen ID/AMR Enrichment Kit Set C (RUO) (96 indexes, 96 samples)
20090310
Includes all reagents necessary for library prep and enrichment including tagementation and PCR reagents, IDT for Illumina DNA/RNA UD Indexes Set C, and Urinary Pathogen ID/AMR Panel to prepare libraries from 96 samples followed by 32, 3-plex enrichment reactions.
List Price:
Discounts:
Urinary Pathogen ID/AMR Enrichment Kit Set D (RUO) (96 indexes, 96 samples)
20090311
Includes all reagents necessary for library prep and enrichment including tagementation and PCR reagents, IDT for Illumina DNA/RNA UD Indexes Set D, and Urinary Pathogen ID/AMR Panel to prepare libraries from 96 samples followed by 32, 3-plex enrichment reactions.
List Price:
Discounts:
Showing of
Product
Qty
Unit Price
Product
Catalog ID
Quantity
Unit price
No, culturing the sample before DNA extraction is not necessary. The Urinary Pathogen ID/AMR Enrichment Kit contains all library prep and enrichment reagents, indexes, and panel probes for on-bead tagmentation followed by a single hybridization step providing a rapid workflow for generating enriched libraries for sequencing.
We recommend isolating DNA using spin-column kits, as these are the most reliable and are commonly used for NGS. The DNA extraction protocol for the Urinary Pathogen ID/AMR Enrichment Kit can be found here.
The library preparation and sequencing steps can be completed in two days on benchtop sequencing systems regardless of pathogen type. However, detection can be completed in < 24 hours when using the MiniSeq Sequencing System and MiniSeq Rapid Reagent Kit during the sequencing process.
The panel in this kit includes probes that will recognize 10 different commercially available controls. Those controls, however, are not provided as part of this kit and must be purchased separately and then spiked into samples prior to DNA and RNA extraction. Only one spike-in control is necessary for each experiment/sample.
We generally recommend starting with 1M single reads, or 2M paired-end reads per sample using a minimum of 1 × 100 bp or 2 × 100 bp sequencing chemistries. These numbers are just a starting point and should be optimized based on sample types and target abundance.
Yes. If the targets covered in this panel align with your experimental goals, you can use DNA extracted from a variety of sample types. Wastewater samples are often tested using this kit for surveillance of these pathogens and their associated AMR. For wastewater samples, we generally recommend increasing the number of reads to 10M single reads or 20M paired-end reads.
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.