Cancer research applications
Explore the different applications within cancer research, including liquid biopsy research, single-cell analysis, and epigenetics.
Reaching a better understanding of tumor immunogenicity and immunotherapy response
Leading immuno-oncology researchers are leveraging next-generation sequencing (NGS) to study immunotherapy response factors, biomarkers, and genomics to improve personalized immunotherapy research.
Immuno-oncology research into the mechanisms tumors use to evade the immune response has led to promising therapeutic targets. These therapies boost the immune system's ability to target cancer or limit the tumor’s ability to evade the immune response. In addition, NGS can help identify which pathways are activated in the tumor environment and how they are involved in processes such as cancer cell proliferation, survival, invasion, and metastasis.

Our understanding of the complex pathways used by cancer cells to evade immune detection continues to expand rapidly. Leading researchers are leveraging NGS technologies to discover biomarkers and develop analytical tools that will guide future development of personalized immunotherapeutics. The key to this approach is to identify the appropriate neoantigen-reactive T-cells that can mediate a durable antitumor immune response.1
NGS is a powerful tool that can provide a detailed analysis of the cancer genome. It can also efficiently assess tumor microenvironments in real-time with high sensitivity to monitor immune marker expression in response to tumor growth or treatment. NGS can characterize the immune cell repertoire, identify various cell populations in the microenvironment, and comprehensively quantify gene expression in thousands of targets simultaneously.
NGS can also help researchers identify neoantigens, study innovative therapies to boost the immune response, and understand how genetic variation can influence their efficacy. NGS has also enabled the predictive selection of novel tumor antigens that can be applied to elicit a tumor-specific response.
Spatial transcriptomics provides a topographical arrangement of gene expression patterns mapped onto tissue sections to link structure and activity. This capability allows researchers to articulate biological interactions at the cellular level to gain novel insights into complex tissues, such as tumor microenvironments.
NGS can be used to study the influence of host-microbiome interactions on cancer development, progression, and treatment efficacy. NGS can profile microbial communities in different contexts, which is critical for identifying species or conditions that may be targeted for developing new therapeutic approaches to cancer.
Single-cell RNA sequencing (scRNA-Seq) is being increasingly used to investigate the transcriptomic profile of cancers and tumor microenvironments at single-cell resolution. Immune cell studies, in combination with single-cell transcriptomics, has been applied to understand the key factors in cancer immunosurveillance and resistance to immunotherapy.
Immune repertoire sequencing (IR-Seq) uses NGS to profile the B cell and T cell receptor repertoire. IR-Seq allows scientists to study lymphocyte receptor variants in various cancers, perform hematological malignancy research, and more.
This application note highlights recent advances in immuno-oncology, including evolving trends, needs of researchers, and genomic technologies available to aid in this rapidly advancing field.
NGS methods allow researchers to study key alterations in immune checkpoint expression, tumor mutational burden (TMB), tumor microenvironment, and gut microbial composition, with high analytical sensitivity.
checkmark
|
Key application |
| Neoantigen, mutational burden | Expression profiling | Microbiome (16S) sequencing | TCR/BCRa profiling | Epigenetic profiling | |
|---|---|---|---|---|---|
| Therapeutic applications | |||||
| Checkpoint inhibitors | checkmark
|
checkmark
|
checkmark
|
checkmark
|
|
| Vaccines | checkmark
|
checkmark
|
checkmark
|
checkmark
|
|
| Adoptive cell therapy | checkmark
|
checkmark
|
checkmark
|
checkmark
|
|
| Prognostics | |||||
| Microbiome | checkmark
|
||||
| Immune repertoire | checkmark
|
checkmark
|
checkmark
|
||
| Cancer monitoring | checkmark
|
checkmark
|
checkmark
|
Explore the different applications within cancer research, including liquid biopsy research, single-cell analysis, and epigenetics.
Learn about NGS-based cancer sequencing methods, which provide more information in less time than single-gene and array-based approaches.

The Cancer Research Methods Guide is a 40+ page comprehensive resource with simple, comprehensive workflows for a broad range of cancer research applications. This guide includes single-cell sequencing, spatial sequencing, methylation profiling, multiomics, cell-free RNA sequencing, and more.
Your email address is never shared with third parties.
Multiomic studies integrate high-dimensional data sets from genomic, epigenomic, transcriptomic, metagenomic, and proteomic approaches, often using computational and network biology to interpret the vast amounts of data generated by these techniques. Applying multiomics to clinical research data amplifies the discovery power of existing omics methodologies to uncover new biomarkers and immunotherapy targets.
In addition, multiomic approaches can provide a comprehensive view of the molecular profile of the tumor and microenvironment. Multiple layers of information provide novel insights into tumor biology that cannot be resolved by single omics studies alone.

Illumina offers several library preparation and sequencing options with access to data analysis options for immuno-oncology research. Streamlined workflows and flexible kit configurations can accommodate multiple study designs.

Watch how innovations in AI and informatics are simplifying the complexity of NGS, making clinical research accessible to a wider range of labs.
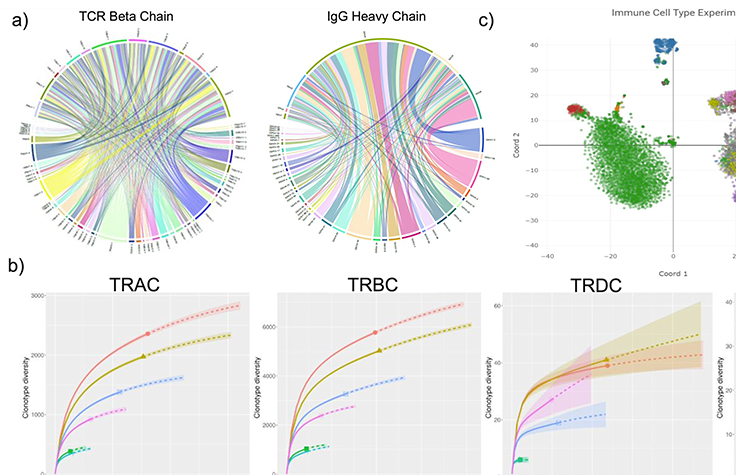
This downloadable poster outlines a new scalable tool for cancer research to enable full-length V(D)J immune-repertoire sequencing (IR-Seq).

Our sequencing and microarray technologies support a broad range of cancer genomics research topics, from DNA to RNA analysis, epigenetics, and more.
Interested in learning more about immuno-oncology research methods?
Your email address is never shared with third parties.