Next-Generation Sequencing (NGS)

Next-generation sequencing for beginners
We'll guide you through the basics of NGS, with tutorials and tips for planning your first experiment.
Get startedWhat is next-generation sequencing?
Next-generation sequencing (NGS) is a massively parallel sequencing technology that offers ultra-high throughput, scalability, and speed. The technology is used to determine the order of nucleotides in entire genomes or targeted regions of DNA or RNA. NGS has revolutionized the biological sciences, allowing labs to perform a wide variety of applications and study biological systems at a level never before possible.
Today's complex genomics questions demand a depth of information beyond the capacity of traditional DNA sequencing technologies. NGS has filled that gap and become an everyday tool to address these questions.

Applications of NGS
Next-generation sequencing technology has fundamentally changed the kinds of questions scientists can ask and answer. Innovative sample preparation and data analysis options enable a broad range of applications. For example, NGS allows labs to:
- Rapidly sequence whole genomes
- Deeply sequence target regions
- Utilize RNA sequencing (RNA-Seq) to discover novel RNA variants and splice sites, or quantify mRNAs for gene expression analysis
- Analyze epigenetic factors such as genome-wide DNA methylation and DNA-protein interactions
- Sequence cancer samples to study rare somatic variants, tumor subclones, and more
- Study the human microbiome
- Identify novel pathogens
How does Illumina NGS work?
Illumina NGS technology utilizes a fundamentally different approach from the classic Sanger chain-termination method. It leverages sequencing by synthesis (SBS) chemistry – tracking the addition of labeled nucleotides as the DNA chain is copied – in a massively parallel fashion.
Next-generation sequencing technology generates masses of DNA sequencing data, and is both less expensive and less time-consuming than traditional Sanger sequencing.1,2 Illumina sequencing systems can deliver data output ranging from 300 kilobases up to multiple terabases in a single run, depending on instrument type and configuration.

In-depth NGS introduction
This detailed overview of Illumina sequencing describes the evolution of genomic science, major advances in sequencing technology, key methods, the basics of Illumina sequencing chemistry, and more.
Read introductionNGS workflow
The next-generation sequencing workflow includes three basic steps: library preparation, sequencing, and data analysis.
Library preparation
NGS library prep education
Learn how the library preparation process works and explore breakthrough technologies to help you get answers quickly.
Library preparation kits
Find kits that support a large range of throughput needs and sample types while boosting workflow efficiency.
Sequencing
NGS platforms
Explore benchtop and production-scale instruments designed to help you choose the right platform for your needs.
Data analysis
NGS data analysis tools
User-friendly, intuitive tools simplify sequencing data analysis, allowing you to focus on research and spend less time configuring workflows.
Methods Guide
Access the information you need—from BeadChips to library preparation for genome, transcriptome, or epigenome studies to sequencer selection, analysis, and support—all in one place. Select the best tools for your lab with our comprehensive guide designed specifically for research applications.
Access guide
Key benefits of NGS
Accessible whole-genome sequencing
Using capillary electrophoresis-based Sanger sequencing, the Human Genome Project took over 10 years and cost nearly $3 billion.
Next-generation sequencing, in contrast, makes large-scale whole-genome sequencing (WGS) accessible and practical for the average researcher. It enables scientists to analyze the entire human genome in a single sequencing experiment, or sequence thousands to tens of thousands of genomes in one year.
Broad dynamic range for expression profiling
NGS-based RNA-Seq is a powerful method that enables researchers to break through the inefficiency and expense of legacy technologies such as microarrays. Microarray gene expression measurement is limited by noise at the low end and signal saturation at the high end.
In contrast, next-generation sequencing quantifies discrete, digital sequencing read counts, offering a broader dynamic range.3,4,5
Tunable resolution for targeted NGS
Targeted sequencing allows you to sequence a subset of genes or specific genomic regions of interest, efficiently and cost-effectively focusing the power of NGS. NGS is highly scalable, allowing you to tune the level of resolution to meet experimental needs. Choose whether to do a shallow scan across multiple samples, or sequence at greater depth with fewer samples to find rare variants in a given region.
Learn more about:
Advances in NGS technology
Recent Illumina next-generation sequencing technology breakthroughs include:
- XLEAP-SBS chemistry: This innovation delivers increased speed and greater fidelity compared to standard Illumina sequencing by synthesis (SBS) chemistry.
- Up to 16 Tb: The NovaSeq X Series provides extraordinary sequencing power to fuel data-intensive applications.
- Breakthrough speed and simplicity: The MiSeq i100 Series offers benchtop sequencing run times as fast as four hours, with streamlined operations and intuitive onboard data analysis.
- Semiconductor sequencing: This technology combines a complementary metal-oxide semiconductor (CMOS) chip with SBS to deliver high-accuracy data in a compact instrument.
- Patterned flow cell technology: This advance provides an exceptional level of throughput for diverse sequencing applications.
Illumina technology innovations
Learn about innovative R&D technologies that are being developed into products, including revolutionary NGS and multiomics technologies.
View innovation roadmapHow scientists use NGS
See how researchers in different fields utilize next-generation sequencing to make breakthrough discoveries.
Advancing drug discovery with microbiome studies
NGS-based whole-genome shotgun sequencing and transcriptomics provide researchers and pharmaceutical companies with data to refine drug discovery and development.
Exploring the tumor microenvironment
Scientists use single-cell NGS techniques to study cancer microenvironments, elucidate gene expression patterns, and gain insights into drug resistance and metastasis.
Cell-free RNA as a noninvasive biomarker
This research highlights the broad potential of circulating cell-free RNA sequencing for biomarker discovery and noninvasive health monitoring.
˝
Expand your research with multiomics
Combine data from genomics, transcriptomics, epigenetics, and/or proteomics to achieve a more comprehensive understanding of molecular changes contributing to disease, cellular response, and development. Perform multiomic experiments using NGS to identify biomarkers, connect genotype to phenotype, and more.
Learn more about multiomics
Start Using NGS
System Buying Guidance
The resources below offer valuable guidance to scientists who are considering purchasing a next-generation sequencing system.
- View NGS platform comparison tables
- Use our interactive platform selection tool
- Read our NGS System Buyer's Guide to determine what factors to consider before making your purchase.
NGS Experimental Considerations
Learn about read length, coverage, quality scores, and other experimental considerations to help you plan your sequencing run.
Use our interactive tools to help you create a custom NGS protocol or select the right products and methods for your project.

Resources for NGS labs
- Sequencing methods: Explore a broad range of methods, from whole-genome sequencing to mRNA-Seq, exome sequencing, single-cell sequencing, and more.
- High-throughput sequencing: Process more samples to improve statistical power. Cost-effectively run data-rich applications using the latest large-scale sequencers.
- Library prep automation: Explore automated liquid handling solutions designed to help labs prepare large quantities of NGS libraries.
- NGS data storage: Securely store vast quantities of NGS and other genomics data.

Proven benchtop sequencing solutions
Illumina offers a proven track record of benchtop sequencing solutions that empower scientists to advance and accelerate their research.
Learn moreSee all benchtop sequencers
Interested in learning more about how to perform NGS? Sign up to have a sales rep contact you.
Featured Webinars
Single cell multiomics: RNA-Seq and beyond
Dr Michael Kelly from the National Cancer Institute discusses use of single-cell and spatial technologies across both basic science and translational research.
Development of XLEAP-SBS chemistry
The Illumina R&D team showcases the work that went into building XLEAP-SBS chemistry in this on-demand webinar.
NGS in microbiology clinical research
Dr Arryn Craney discusses NGS implementation options for microbiology clinical research labs and hands-on experience with respiratory pathogen sequencing.
Genomics news

The next generation is shaping the future of rare diseases
Youth advocates such as Liam McCarthy are ensuring their voices are included in global conversations
Read article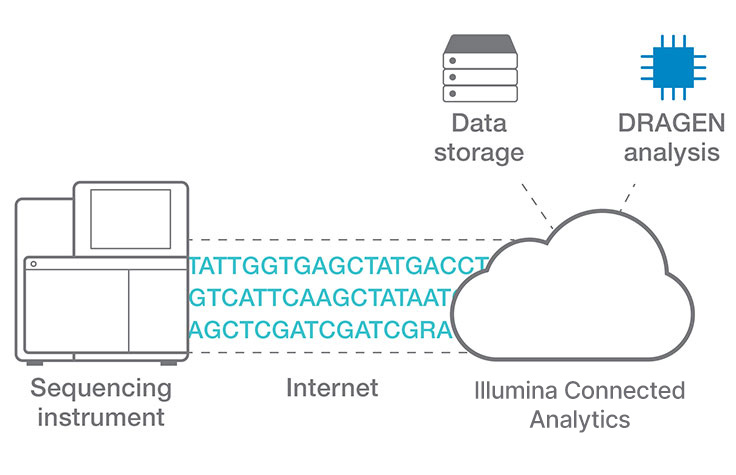
Efficient cloud data analysis for COPD multiomics project
Researchers at Okayama University in Japan use Illumina Connected Analytics with DRAGEN pipelines for analyzing whole-genome, exome, transcriptome, and metagenome data
Read Interview
The future of global food security may depend on quality agrigenomics software
In Hasselt, Belgium, Breed Bio’s next-level LIMS is reimagining the user experience and improving agriculture
Read articleAdditional resources
RNA sequencing (RNA-Seq)
RNA-Seq uses next-generation sequencing to analyze expression across the transcriptome, enabling scientists to detect known or novel features and quantify RNA.
Sequencing services
Access fast, reliable next-generation sequencing services that provide high-quality data and offer extensive scientific expertise.
Illumina NGS & microarray training
Work with expert Illumina instructors and get hands-on training. We also offer online courses, webinars, videos, and podcasts.
References
- Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate Whole Human Genome Sequencing using Reversible Terminator Chemistry. Nature. 2008; 456 (7218): 53–59.
- Shendure J and Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008; 26(10): 1135-1145.
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009; 10: 57–63.
- Wilhelm BT, Landry JR. RNA-Seq—quantitative measurement of expression through massively parallel RNA sequencing. Methods. 2009; 48: 249–57.
- Zhao S, Fung-Leung WP, Bittner A, and Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014; 16;9(1): e78644.
