Index Hopping
Minimize index hopping in multiplexed runs
Tips and best practices to avoid sequencing read misalignment associated with index switching
What Is Index Hopping?
Index hopping or index switching is a known phenomenon that has impacted NGS technologies from the time sample multiplexing was developed.1 It causes a specific type of misassignment that results in the incorrect assignment of libraries from the expected index to a different index in a multiplexed pool.
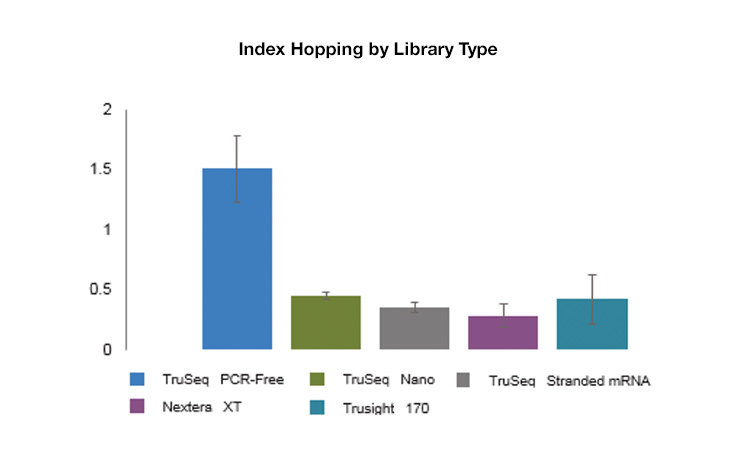
Index hopping is a rare occurrence, but it can be seen at slightly elevated levels on instruments using patterned flow cells with exclusion amplification chemistry versus those that do not use patterned flow cells. Typical levels of index hopping on patterned flow cell systems range from 0.1–2%, depending on the type, quality, and handling of the library.
Index Hopping Effects and Mitigation Strategies
Gain insights on typical levels of index hopping and the downstream effect on applications.
View WebinarIncreasing Capacity with Multiplexing
Improvements in next-generation sequencing (NGS) technology have greatly increased sequencing speed and data output, resulting in the massive sample throughput of current sequencing platforms. A key to utilizing this increased capacity is multiplexing, which adds unique sequences, called indexes, to each DNA fragment during library preparation. This allows large numbers of libraries to be pooled and sequenced simultaneously during a single sequencing run.
Gains in throughput from multiplexing come with an added layer of complexity, as sequencing reads from pooled libraries need to be identified and sorted computationally in a process called demultiplexing before final data analysis. However, with multiplexing, the potential for index hopping is present regardless of the library prep method or sequencing system used. Index hopping may result in assignment of sequencing reads to the wrong index during demultiplexing, leading to misalignment.

Effects of Index Misassignment
Learn why it happens and best practices to reduce the impact of index hopping.
Read White PaperWhat is the Impact of Index Hopping?
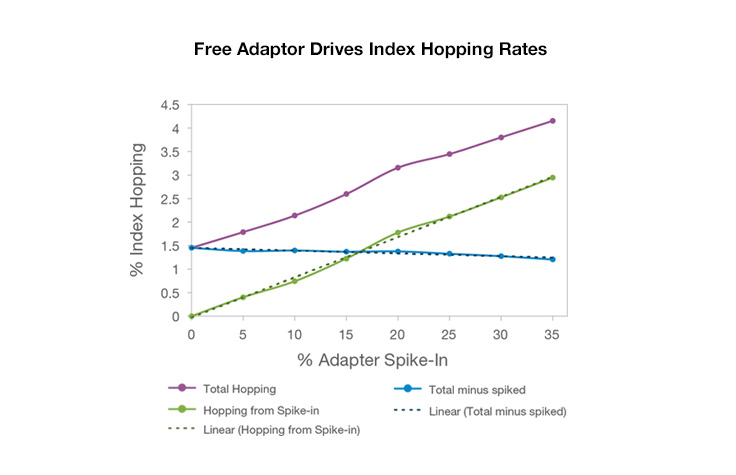
While index hopping can occur, and can be seen at slightly elevated levels on instruments that use patterned flow cells, it has a limited effect on most applications, and background hopped reads can be filtered out as noise. Libraries with higher levels of free adapters will see higher levels of index hopping. However, use of unique dual indexing combinations eliminates hopped reads from downstream analysis because unexpected combinations will be assigned as undetermined.
How to Minimize Index Hopping
To minimize the level and effect of index hopping, customers should follow these library preparation tips and best practices:
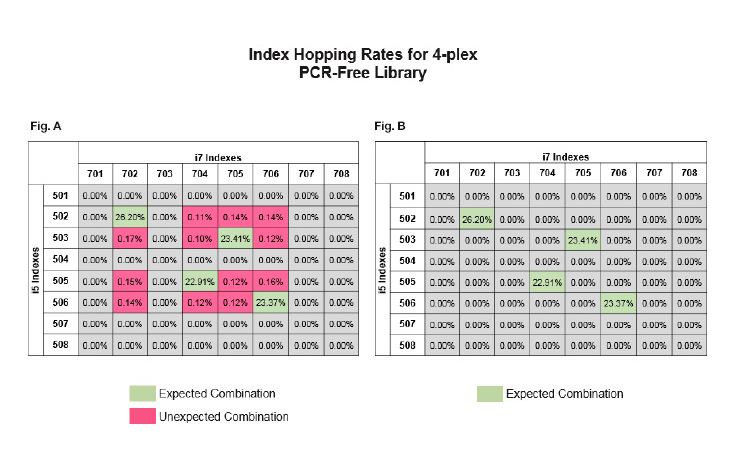
- Use unique dual indexing (UDI) pooling combinations (unique i5 and i7 indexes)
- Remove free adapters from library preps
- Store libraries individually at -20°C
- Pool libraries prior to sequencing
Dual Index Sequencing
When pooling libraries for sequencing, we recommend using unique dual indexes over combinatorial dual indexes. Unique dual indexing mitigates index hopping by filtering hopped reads.
Learn More About Unique Dual IndexingUnique Molecular Identifiers
Unique molecular identifiers (UMIs) are a type of molecular barcoding that provides error correction and increased accuracy during sequencing. Sequencing with UMIs can reduce the rate of false-positive variant calls and increase sensitivity of variant detection.
Learn More About UMIsIndex Hopping Mitigations
Unique dual indexes allow researchers to remove unexpected combinations and only focus on the ‘true’ data with correct index combinations. We are expanding the number of unique dual indexes we offer. Examples of library preparation products that are compatible with unique dual indexes include:
Additional Information
For more in-depth information, see the following resources:
- Understanding UDIs & Associated Library Prep Kits (technical bulletin)
- NGS Tutorials (training and videos)
Interested in receiving newsletters, case studies, and information from Illumina based on your area of interest?
Sign up now.
Related Solutions
Patterned Flow Cell Technology
Fixed spacing of sequencing clusters with defined feature sizes contributes to increased data output, reduced costs, and faster run times.
Multiplex Sequencing with Index Adapters
Sample multiplexing with index adapters dramatically increases the number of samples analyzed in a single sequencing run.
Illumina Training
Work with expert Illumina instructors and get hands-on NGS training. We also offer online courses and webinars.
References
- Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012:2513–2524.