Low-Quality/FFPE RNA-Seq Solutions
Enabling RNA-Seq Analysis of FFPE Samples
RNA sequencing (RNA-Seq) analysis of FFPE and other low-quality samples can offer key insights for disease research. Unfortunately, much of this data used to be inaccessible. FFPE RNA-Seq solutions from Illumina now enable researchers to obtain this valuable data, yielding high-quality results from challenging samples.
Considerations for RNA-Seq Read Length and Coverage
This bulletin reviews RNA sequencing considerations and offers resources for planning RNA-Seq experiments.
Read Illumina knowledgemRNA-Seq for FFPE and Low-Quality Samples
mRNA sequencing (mRNA-Seq) is an accurate and cost-effective method for analyzing the coding transcriptome. Focusing your sequencing budget on only the messenger RNAs allows for increased sample throughput and lower per sample costs relative to total RNA-Seq, while enabling discovery of novel features such as alternative splicing, fusion transcripts and coding variants.1,2
RNA Exome Capture Sequencing overcomes the limitation of traditional poly-A-based methods for capturing the coding transcriptome; these traditional methods are ineffective for FFPE RNA-Seq analysis. By applying sequence-specific capture that does not rely on the presence of polyadenylated transcripts, this method is ideal for mRNA-Seq with FFPE or degraded samples and samples with limited starting material.
Featured RNA-Seq Research

Deciphering the Role of Long Non-Coding RNA in Cancer
Researchers use RNA-Seq to study lncRNAs and explore their potential as cancer biomarkers.
Read Interview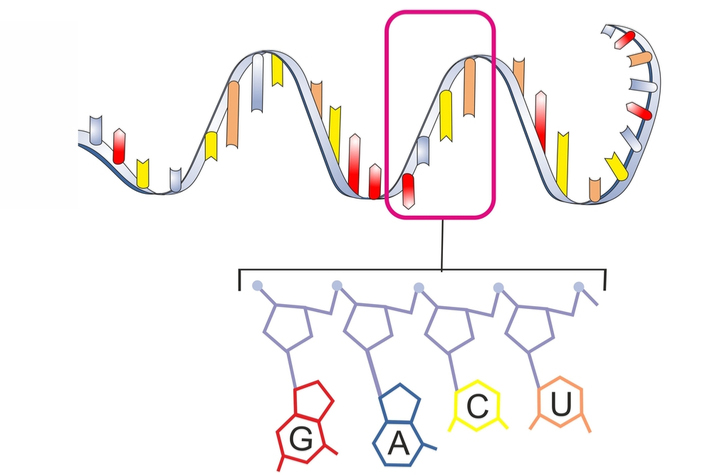
A Tale of Two RNA Library Prep Kits
A critical comparison between two popular library prep kits reveals information of interest to researchers conducting RNA sequencing studies.
Read Interview
RNA Sequencing Methods Review
This collection contains protocol diagrams, advantages and disadvantages, and peer-reviewed RNA-Seq publications.
Read ReviewTotal RNA-Seq for FFPE and Low-Quality Samples
Total RNA sequencing provides a comprehensive view of the transcriptome, enabling analysis of both coding and multiple forms of noncoding RNA in a single experiment.
This method provides highly accurate transcript abundance information, and can detect novel features such as gene fusions, transcript isoforms, cSNPs, and allele-specific expression.
QC Recommendations for FFPE RNA-Seq
Extraction methods for FFPE tissue generally yield highly degraded RNA. Read our FFPE quality control recommendations to determine whether your FFPE samples are viable input material for Illumina library preparation kits.
View Tips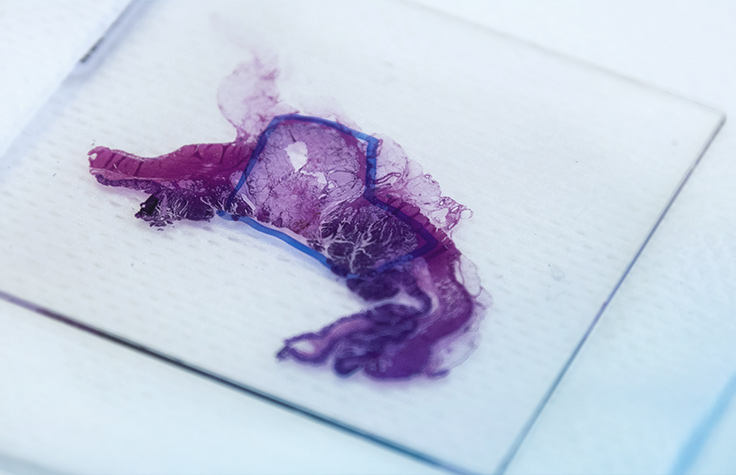
Targeted RNA Sequencing
Targeted RNA sequencing is a highly accurate and specific method for measuring expression of transcripts of interest, offering researchers both quantitative and qualitative information. This allows for differential expression analysis, as well as allele-specific expression measurement and verification of the presence of fusion genes.
Targeted cancer sequencing with FFPE-compatible RNA panels allows researchers to study gene expression changes and gene fusions, providing a focused view of the functionally relevant changes occurring in cancer.
Interested in receiving newsletters, case studies, and information from Illumina based on your area of interest? Sign up now.
Related Links
Library Prep and Array Kit Selector
Determine the best kit for your needs based on your project type, starting material, and method of interest.
Cancer RNA-Seq
Sequencing the coding regions or the whole cancer transcriptome can provide researchers with valuable information about gene expression changes in tumors.
Easily Analyze RNA-Seq Data
User-friendly tools simplify data analysis for the most common RNA sequencing study designs.
References
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63.
- Wilhelm BT, Landry JR. RNA-Seq—quantitative measurement of expression through massively parallel RNA sequencing. Methods. 2009;48:249–57.