mRNA Sequencing
Introduction to mRNA Sequencing
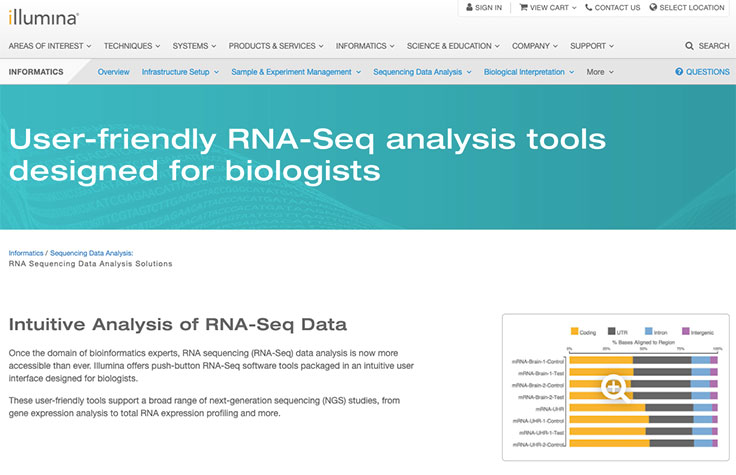
mRNA sequencing (mRNA-Seq) has rapidly become the method of choice for analyzing the transcriptomes of disease states, of biological processes, and across a wide range of study designs. In addition to being a highly sensitive and accurate means of quantifying gene expression, mRNA-Seq can identify both known and novel transcript isoforms, gene fusions, and other features as well as allele-specific expression. Next-generation sequencing (NGS)-based mRNA-Seq delivers a comprehensive view of the coding transcriptome that is not restricted by the filter of prior knowledge.
Sifting Through Cells
Dr. Norma Neff discusses how researchers at Stanford University use single-cell mRNA-Seq to understand early development.
Read interviewAdvantages of mRNA Sequencing
mRNA-Seq provides a number of advantages over gene expression arrays in analyzing the transcriptome.
- Offers a broader dynamic range, enabling more sensitive and accurate measurement of fold changes in gene expression
- Captures both known and novel features
- Can be applied across a wide range of species
Transitioning from Arrays to mRNA-Seq
Expression Analysis developed tools to make it easier to compare mRNA-Seq results with previous array data.
Read InterviewAccurate, High-Resolution View of the Transcriptome
Ratio compression is an established technical limitation of gene expression arrays that reduces dynamic range and can mask or alter measured transcriptional changes.1–3 In contrast, mRNA-Seq is not subject to this bias and provides more comprehensive and accurate measurements of gene expression changes.
Additionally, mRNA-Seq can provide strand information, which enables the detection of antisense expression, allows more accurate quantification of overlapping transcripts, and increases the percentage of alignable reads.
Autism and mRNA-Seq
Stanley Lapidus, President, CEO, and Founder, SynapDx, discusses how the company is using mRNA-Seq to study autism.
View VideoRecommended mRNA-Seq Workflow for Standard Samples
Library Prep
Illumina Stranded mRNA Prep
A simple, scalable, cost-effective, rapid single-day solution for analyzing the coding transcriptome leveraging as little as 25 ng input of standard (non-degraded) RNA.
Sequencing
NextSeq 1000 & 2000 Systems
These cost-efficient, user-friendly, mid-throughput benchtop sequencers support mRNA-Seq plus a wide variety of other current and emerging applications.
MiSeq i100 Series
Our fastest, simplest benchtop system for mRNA sequencing.
Data Analysis
DRAGEN RNA Pipeline
Performs alignment, quantification, and fusion detection.
RNA-Seq Differential Expression
Enables differential gene expression analysis.

RNA sequencing methods guide
This guide provides Illumina solutions for profiling RNA, from targeted panels to mRNA to the whole transcriptome. Illumina RNA sequencing workflows seamlessly integrate library prep, sequencing, and data analysis to support transcriptome research.
Download guideRNA Library Preparation
Advances in RNA-Seq library prep are revolutionizing the study of the transcriptome. Our enhanced RNA sequencing library prep portfolio spans multiple types of sequencing studies, from mRNA-Seq to total RNA-Seq and more. These solutions offer rapid turnaround time, broad study flexibility, and sequencing scalability.
Learn more about RNA library prepComprehensive mRNA-Seq Workflow
Illumina sequencing by synthesis (SBS) chemistry is the most widely adopted NGS technology, producing approximately 90% of global sequencing data.*
In addition to our industry-leading data quality, Illumina offers integrated mRNA-Seq workflows that simplify the entire process, from library preparation to data analysis and biological interpretation.
Featured mRNA Sequencing Articles

Mapping Neural Diversity
Allen Institute researchers use mRNA-Seq to analyze gene expression in individual neurons and classify V1 neural cells.
Read Interview
Single-Cell Analysis in Developmental Biology
Dr. Colin Trapnell discusses his lab's experience with single-cell mRNA-Seq and his efforts to make bioinformatics tools available to all.
Read Interview
RNA Sequencing Considerations
Learn about read length and depth requirements for RNA-Seq and find resources to help with experimental design.
Read Illumina Knowledge ArticleRelated Solutions
RNA-Seq in Cancer Research
Monitoring gene expression changes with mRNA-Seq can help researchers identify biomarkers predictive of disease prognosis or response to therapy. Learn more about cancer RNA-Seq.

Gene Expression Analysis for Disease Studies
RNA-Seq-based gene expression profiling studies can provide visibility into how genetic and environmental factors contribute to a broad range of diseases. Learn more about gene expression profiling.

RNA Drug Response Biomarker Discovery
Find out how to utilize RNA-Seq to identify novel RNA-based drug response biomarkers. Access resources designed to help new users adopt this application. Learn more about drug response RNA biomarker analysis.
Interested in receiving newsletters, case studies, and information on sequencing methods?
Enter your email address.
Additional Resources
References
- Shi L, Tong W, Su Z, et al. Microarray scanner calibration curves: characteristics and implications. BMC Bioinformatics. 2005;6 Suppl 2:S11.
- Naef F, Socci ND, Magnasco M. A study of accuracy and precisions in oligonucleotide arrays: extracting more signal at large concentrations. Bioinformatics. 2003;19:178-184.
- Yuen T, Wurmbach E, Pfeffer RL, Ebersole BJ, Sealfon SC. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 2002;30:e48.
*Data calculations on file. Illumina, Inc., 2015