Illumina sequencing-by-synthesis (SBS) has significantly advanced genome mapping methods over the past 20 years, helping researchers achieve highly accurate coverage over the vast majority of the human genome.1 However, for a small percentage of the genome, mapping short reads to a reference genome remains challenging. These challenges primarily occur in repetitive or low-complexity regions, regions that have high homology with other parts of the genome, or large structural variations.
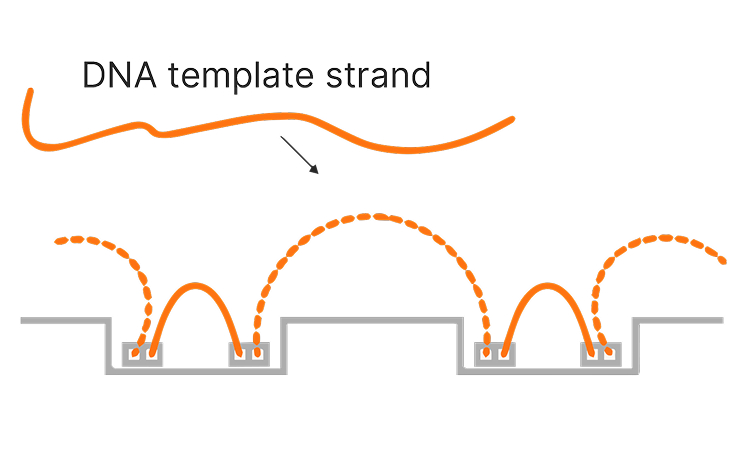
Mapped read technology leverages on-flow cell library preparation and novel informatics that incorporate proximity information from clusters in neighboring nanowells to generate long-range genomic insights. The unique workflow maintains the link between the original long DNA template and the resulting short sequencing reads, enabling improved mapping in low-complexity regions, ultra-long phasing of genetic variants, and enhanced detection of structural variants.